Note
This page was generated from
3D Reconstruction.ipynb.
Interactive online version:
.
Some tutorial content may look better in light mode.
Three dims models reconstruction#
This notebook demonstrates the process of 3D models reconstruction based on 3D spatial transcriptome data. This is done in the following five steps:
Construct the original 3D point cloud model (Embryo);
Construct the mesh model based on the 3D point cloud model (Embryo);
Construct the cell model based on the 3D point cloud model (Embryo);
Construct the voxel model based on the mesh model (Embryo);
Construct the subtype models based on the embryo model (CNS);
This example reconstructs 3D Drosophila Embryo/CNS models based on 3D spatial transcriptomics data of Drosophila Embryo.
[1]:
import warnings
warnings.filterwarnings('ignore')
import numpy as np
import spateo as st
2023-07-25 09:35:00.085195: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Could not find TensorRT
Load the data#
[2]:
cpo = [(553, 1098, 277), (1.967, -6.90, -2.21), (0, 0, 1)]
adata = st.sample_data.drosophila(filename="E7-9h_cellbin.h5ad")
adata.uns["__type"] = "UMI"
adata
[2]:
AnnData object with n_obs × n_vars = 25921 × 8136
obs: 'area', 'slices', 'anno_cell_type', 'anno_tissue', 'anno_germ_layer', 'actual_stage'
uns: '__type'
obsm: '3d_align_spatial'
layers: 'counts_X', 'spliced', 'unspliced'
3D reconstruction of Drosophila Embryo#
Four types of 3D models can be reconstructed through spateo, including point cloud model, mesh model, cell model and voxel model. - The point cloud model and cell model are used to visualize gene expression matrix, cell annotation results and other information. - The mesh model is used to calculate the morphological characteristics of the 3D model, including the length, width, height, volume, surface area, etc. of the model. - The voxel model is used for gene expression matrix interpolation and smoothing.
In addition to realizing 3D reconstruction through the following code, you can also choose to use spateo-viewer to reconstruct 3D models.
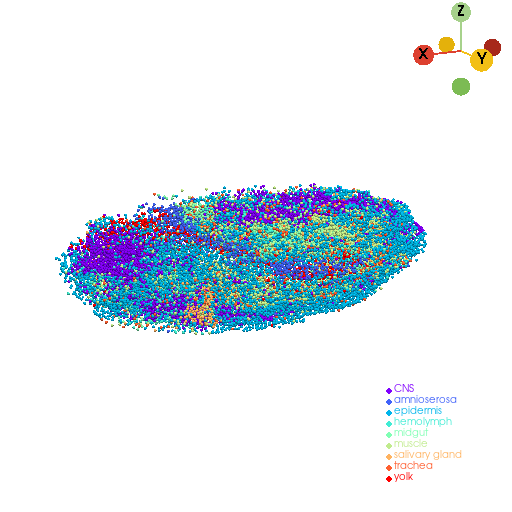
3D reconstruction of original point cloud model#
[3]:
embryo_pc, plot_cmap = st.tdr.construct_pc(adata=adata.copy(), spatial_key="3d_align_spatial", groupby="anno_tissue", key_added="tissue", colormap="rainbow")
st.pl.three_d_plot(model=embryo_pc, key="tissue", model_style="points", colormap=plot_cmap, jupyter="static", cpo=cpo)
st.tdr.save_model(model=embryo_pc, filename="embryo_pc_model.vtk")
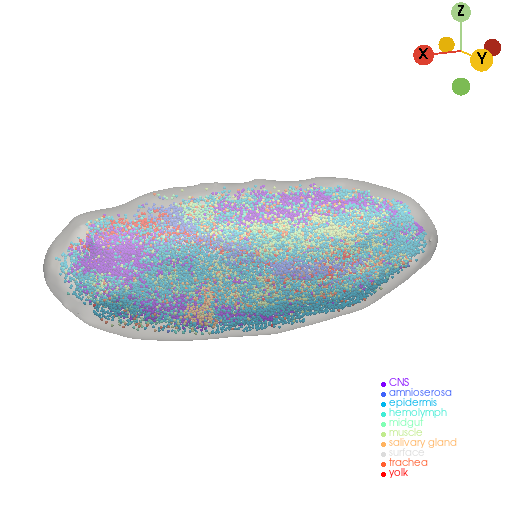
3D reconstruction of mesh model#
[8]:
embryo_mesh, _, _ = st.tdr.construct_surface(pc=embryo_pc, key_added="tissue", alpha=0.6, cs_method="marching_cube", cs_args={"mc_scale_factor": 0.8}, smooth=5000, scale_factor=1.08)
st.pl.three_d_plot(model=st.tdr.collect_models([embryo_mesh, embryo_pc]), key="tissue", model_style=["surface", "points"], jupyter="static", cpo=cpo)
st.tdr.save_model(model=embryo_mesh, filename="embryo_mesh_model.vtk")
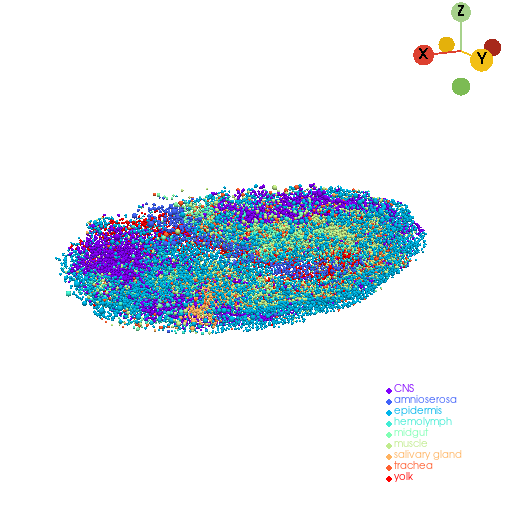
3D reconstruction of cell mesh model#
[9]:
# Add cell radius info
obs_index = embryo_pc.point_data["obs_index"].tolist()
area = adata[obs_index, :].obs["area"].values
cell_radius = pow(np.asarray(area), 1 / 2)
st.tdr.add_model_labels(model=embryo_pc, labels=cell_radius, key_added="cell_radius", where="point_data", colormap="hot_r", inplace=True)
embryo_cells = st.tdr.construct_cells(pc=embryo_pc, cell_size=embryo_pc.point_data["cell_radius"], geometry="sphere", factor=0.2)
st.pl.three_d_plot(model=embryo_cells, key="tissue", jupyter="static", cpo=cpo)
st.tdr.save_model(model=embryo_cells, filename="embryo_cells_mesh_model.vtk")
3D reconstruction of voxel model#
[10]:
embryo_voxel, _ = st.tdr.voxelize_mesh(mesh=embryo_mesh, voxel_pc=None, key_added="tissue", label="embryo_voxel", color="gainsboro", smooth=500)
st.pl.three_d_plot(model=embryo_voxel, key="tissue", jupyter="static", cpo=cpo)
st.tdr.save_model(model=embryo_voxel, filename="embryo_voxel_model.vtk")

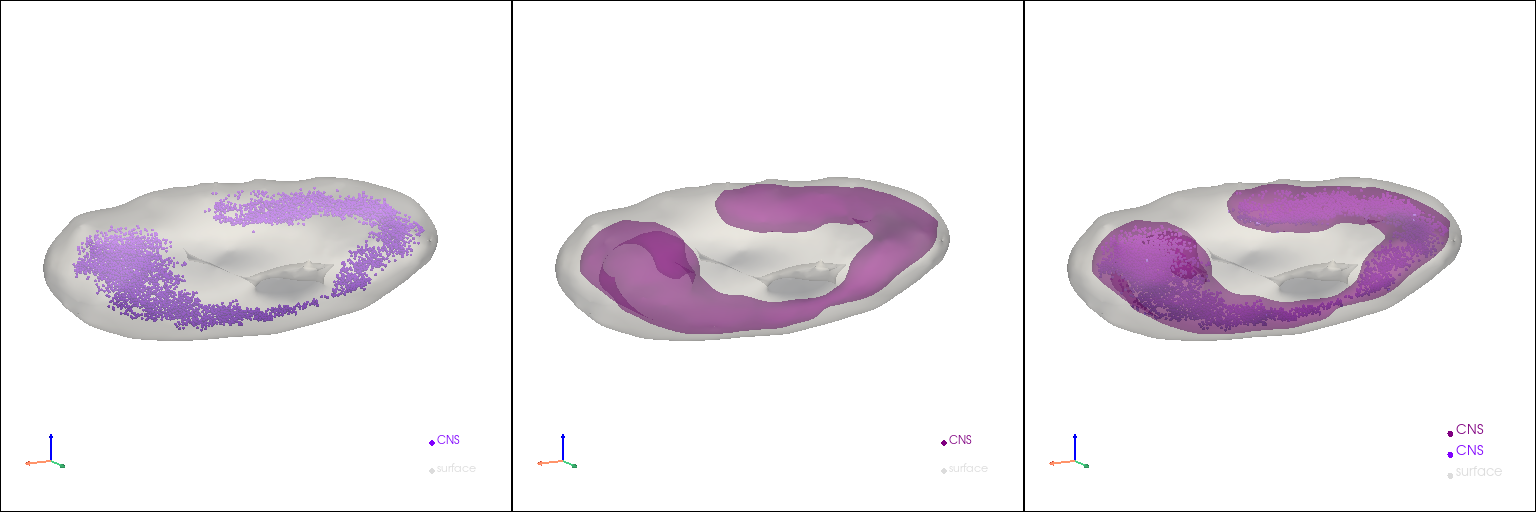
3D reconstruction of subtype models (Drosophila CNS)#
[11]:
subtype = "CNS"
subtype_rpc = st.tdr.three_d_pick(model=embryo_pc, key="tissue", picked_groups=subtype)[0]
subtype_tpc = st.tdr.interactive_rectangle_clip(model=subtype_rpc, key="tissue", invert=True)[0]
[12]:
subtype_mesh, subtype_pc, _ = st.tdr.construct_surface(
pc=subtype_tpc, key_added="tissue", label=subtype, color="purple", alpha=0.6, cs_method="marching_cube", cs_args={"mc_scale_factor": 0.8}, smooth=5000, scale_factor=1
)
[13]:
st.pl.three_d_multi_plot(
model=st.tdr.collect_models(
[
st.tdr.collect_models([embryo_mesh, subtype_pc]),
st.tdr.collect_models([embryo_mesh, subtype_mesh]),
st.tdr.collect_models([embryo_mesh, subtype_mesh, subtype_pc])
]
),
key="tissue",
model_style=[["surface", "points"], "surface", ["surface", "surface", "points"]],
model_size=3,
shape=(1, 3),
jupyter="static",
cpo=[cpo]
)
[ ]: