Note
This page was generated from
Slices Alignment with Morpho.ipynb.
Interactive online version:
.
Some tutorial content may look better in light mode.
Slices Alignment with Morpho#
This notebook demonstrates the process of Aligning spatial transcriptomics data. This is done in the following two steps:
Align spatial transcriptomics data from a set of multi-slices;
Reduce the amount of data by sampling to improve alignment speed.
Here, we develop a novel alignment method, Morpho, which utilized a Gaussian Process algorithm to align and integrate spatial transcriptomics data.
[1]:
import warnings
warnings.filterwarnings('ignore')
import spateo as st
2023-07-24 17:41:40.675118: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Could not find TensorRT
Load the data#
[2]:
# cellbin data
cellbin_slices = st.sample_data.drosophila(filename="E7-9h_cellbin_h5ad.zip")[4:8]
cellbin_slices
[2]:
[AnnData object with n_obs × n_vars = 1745 × 9057
obs: 'area', 'slices'
uns: '__type', 'spatial'
obsm: 'bbox', 'contour', 'spatial',
AnnData object with n_obs × n_vars = 1769 × 9158
obs: 'area', 'slices'
uns: '__type', 'spatial'
obsm: 'bbox', 'contour', 'spatial',
AnnData object with n_obs × n_vars = 1244 × 8992
obs: 'area', 'slices'
uns: '__type', 'spatial'
obsm: 'bbox', 'contour', 'spatial',
AnnData object with n_obs × n_vars = 1715 × 9145
obs: 'area', 'slices'
uns: '__type', 'spatial'
obsm: 'bbox', 'contour', 'spatial']
Align spatial transcriptomics data from a set of multi-slices#
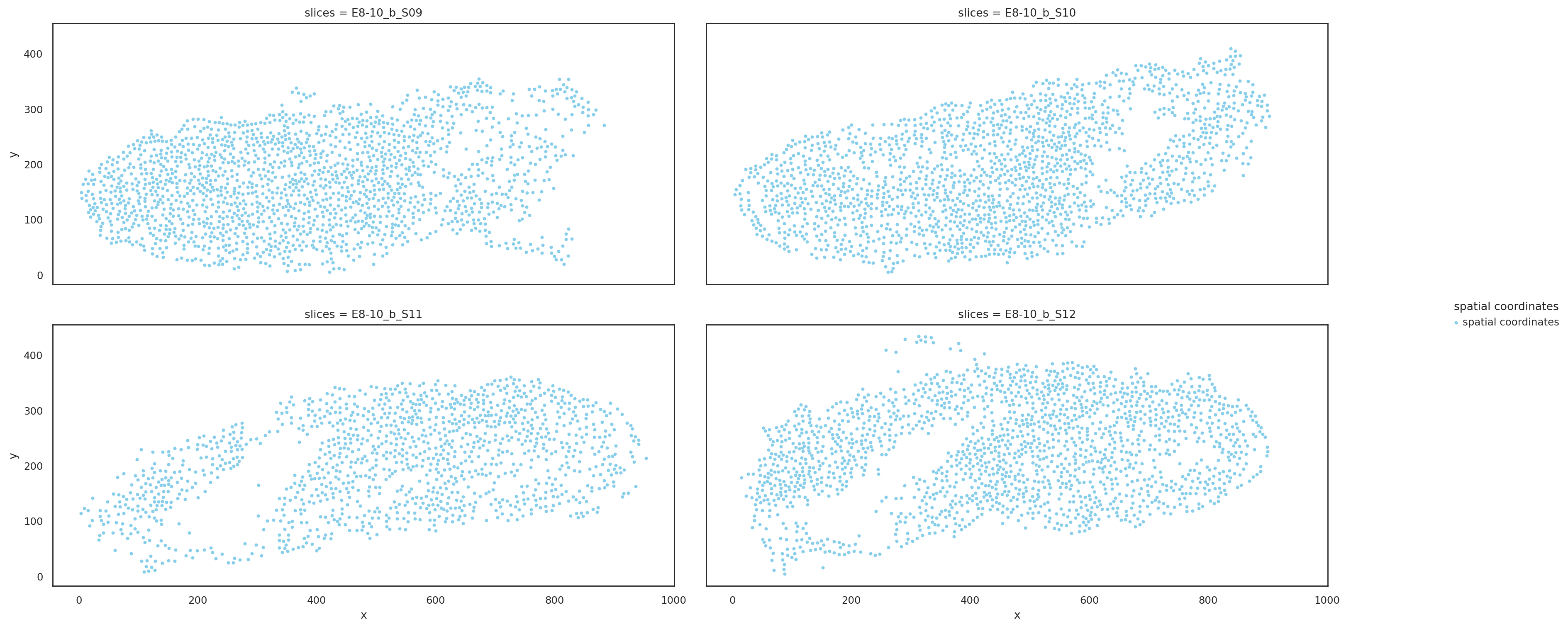
Visualize slices based on raw coordinates#
[3]:
st.pl.multi_slices(
slices=cellbin_slices,
slices_key="slices",
spatial_key="spatial",
point_size=10,
ncols=2,
)
Slices alignment#
[4]:
aligned_slices, pis, sigma2s = st.align.morpho_align(
models=cellbin_slices,
spatial_key="spatial",
key_added="align_spatial",
mode="SN-S",
SVI_mode=True,
device="0" # or device="cpu"
)
|-----> [Models alignment based on morpho, mode: SN-S.] in progress: 33.3333%|-----> Filtered all samples for common genes. There are 8334 common genes.
|-----> Coordinates normalization params:
|-----------> Scale: 232.68878173828125.
|-----> Performing coarse rigid alignment...
|-----> Coarse rigid alignment done.
|-----> [Start morpho alignment] in progress: 100.0000%
|-----> [Start morpho alignment] finished [0.4564s]
|-----> Key Parameters: gamma: 0.9228506088256836; beta2: 0.5335635542869568; sigma2: 0.0030608675442636013
|-----> [Models alignment based on morpho, mode: SN-S.] in progress: 66.6667%|-----> Filtered all samples for common genes. There are 8257 common genes.
|-----> Coordinates normalization params:
|-----------> Scale: 241.8182373046875.
|-----> Performing coarse rigid alignment...
|-----> Coarse rigid alignment done.
|-----> [Start morpho alignment] in progress: 100.0000%
|-----> [Start morpho alignment] finished [0.2478s]
|-----> Key Parameters: gamma: 0.7223951816558838; beta2: 0.5312038660049438; sigma2: 0.0018686357652768493
|-----> [Models alignment based on morpho, mode: SN-S.] in progress: 100.0000%|-----> Filtered all samples for common genes. There are 8251 common genes.
|-----> Coordinates normalization params:
|-----------> Scale: 244.2300567626953.
|-----> Performing coarse rigid alignment...
|-----> Coarse rigid alignment done.
|-----> [Start morpho alignment] in progress: 100.0000%
|-----> [Start morpho alignment] finished [0.2585s]
|-----> Key Parameters: gamma: 0.6794537901878357; beta2: 0.5465249419212341; sigma2: 0.0010000000474974513
|-----> [Models alignment based on morpho, mode: SN-S.] in progress: 100.0000%
|-----> [Models alignment based on morpho, mode: SN-S.] finished [0.0147s]
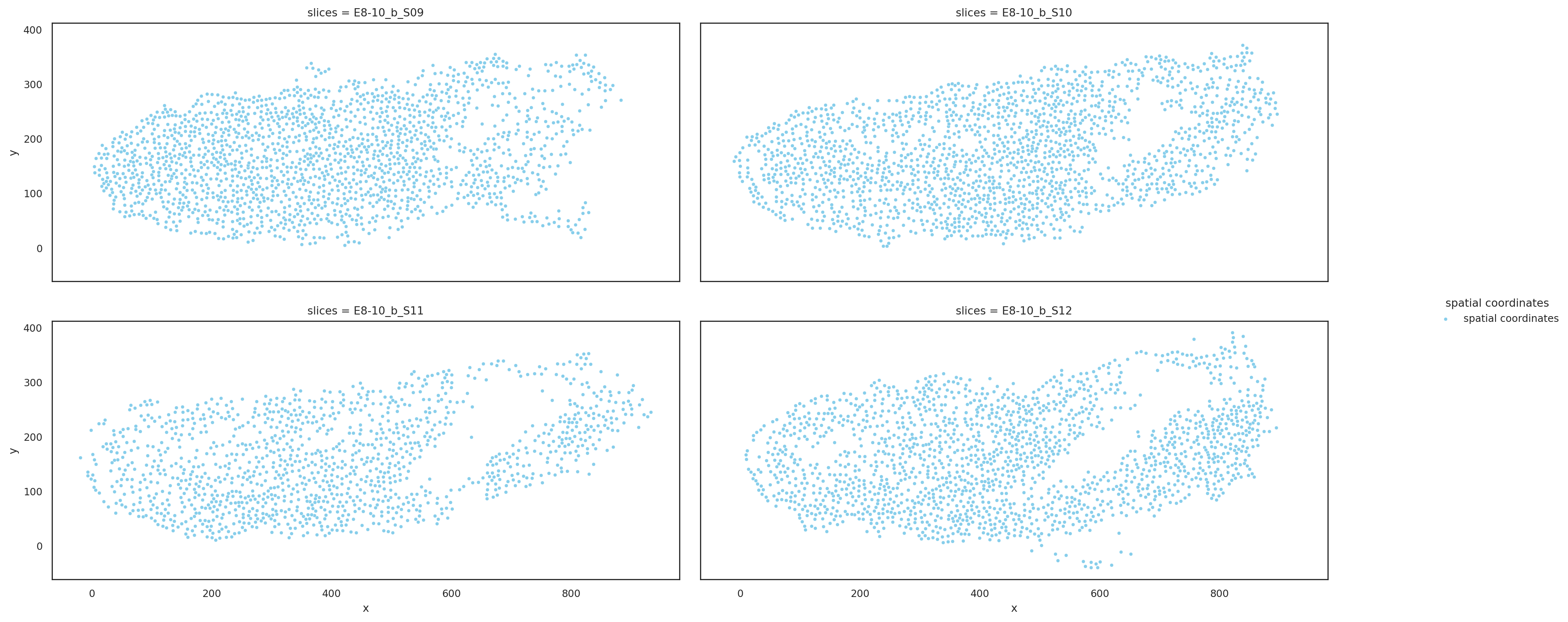
Visualize slices based on aligned coordinates#
[5]:
st.pl.multi_slices(
slices=aligned_slices.copy(),
slices_key="slices",
spatial_key="align_spatial",
point_size=10,
ncols=2,
)
Reduce the amount of data by down-sampling to improve alignment speed#
Slices alignment#
[6]:
aligned_slices, downsampling_slices, pis, pis_downsampling, siaga2s = st.align.morpho_align_ref(
models=cellbin_slices,
models_ref=None,
n_sampling=500,
sampling_method="trn",
spatial_key="spatial",
key_added="align_spatial",
mode="SN-S",
SVI_mode=True,
device="0"
)
|-----> [Running TRN] in progress: 100.0000%|-----> [Running TRN] completed [19.5825s]
|-----> [Running TRN] in progress: 100.0000%|-----> [Running TRN] completed [19.8104s]
|-----> [Running TRN] in progress: 100.0000%|-----> [Running TRN] completed [19.8145s]
|-----> [Running TRN] in progress: 100.0000%|-----> [Running TRN] completed [19.3991s]
|-----> [Models alignment with ref-models based on morpho, mode: SN-S.] in progress: 33.3333%|-----> Filtered all samples for common genes. There are 8334 common genes.
|-----> Coordinates normalization params:
|-----------> Scale: 246.5740203857422.
|-----> Performing coarse rigid alignment...
|-----> Coarse rigid alignment done.
|-----> [Start morpho alignment] in progress: 100.0000%
|-----> [Start morpho alignment] finished [0.2496s]
|-----> Key Parameters: gamma: 0.16604205965995789; beta2: 0.5303851962089539; sigma2: 0.0010000000474974513
|-----> [Models alignment with ref-models based on morpho, mode: SN-S.] in progress: 66.6667%|-----> Filtered all samples for common genes. There are 8257 common genes.
|-----> Coordinates normalization params:
|-----------> Scale: 254.66787719726562.
|-----> Performing coarse rigid alignment...
|-----> Coarse rigid alignment done.
|-----> [Start morpho alignment] in progress: 100.0000%
|-----> [Start morpho alignment] finished [0.4048s]
|-----> Key Parameters: gamma: 0.19782909750938416; beta2: 0.559673011302948; sigma2: 0.0010000000474974513
|-----> [Models alignment with ref-models based on morpho, mode: SN-S.] in progress: 100.0000%|-----> Filtered all samples for common genes. There are 8251 common genes.
|-----> Coordinates normalization params:
|-----------> Scale: 255.73651123046875.
|-----> Performing coarse rigid alignment...
|-----> Coarse rigid alignment done.
|-----> [Start morpho alignment] in progress: 100.0000%
|-----> [Start morpho alignment] finished [0.2497s]
|-----> Key Parameters: gamma: 0.19356141984462738; beta2: 0.5479736328125; sigma2: 0.0010000000474974513
|-----> [Models alignment with ref-models based on morpho, mode: SN-S.] in progress: 100.0000%
|-----> [Models alignment with ref-models based on morpho, mode: SN-S.] finished [0.0051s]
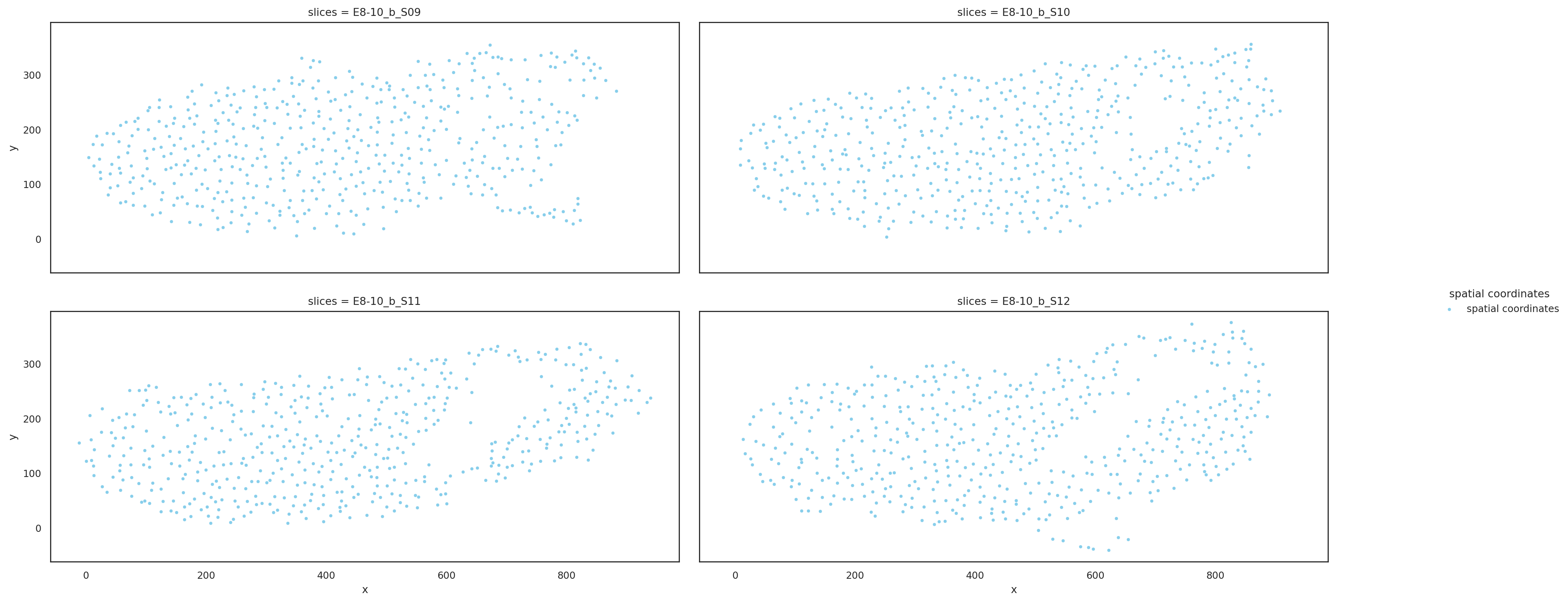
Visualize downsampling slices based on aligned coordinates#
[7]:
st.pl.multi_slices(
slices=downsampling_slices.copy(),
slices_key="slices",
spatial_key="align_spatial",
point_size=10,
ncols=2,
)
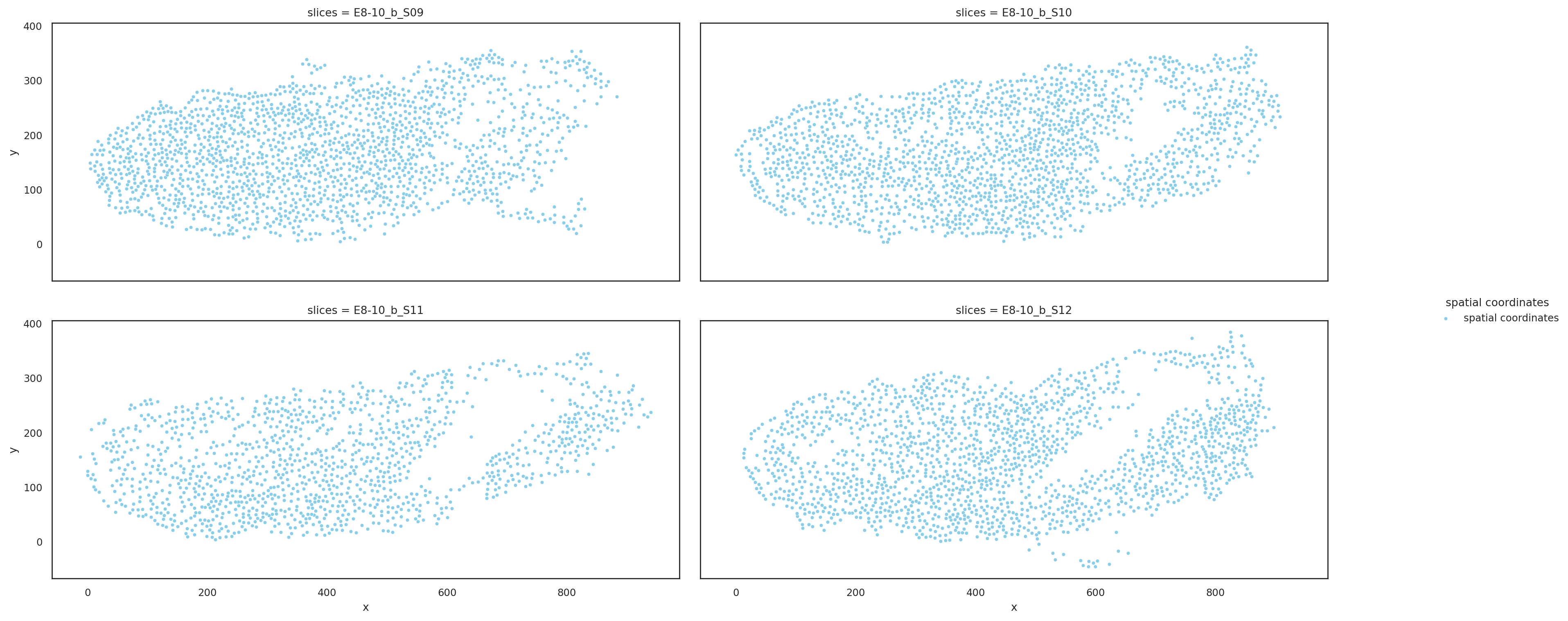
Visualize slices based on aligned coordinates#
[8]:
st.pl.multi_slices(
slices=aligned_slices.copy(),
slices_key="slices",
spatial_key="align_spatial",
point_size=10,
ncols=2,
)
[8]: