Note
This page was generated from
1_models_alignment.ipynb.
Interactive online version:
.
Some tutorial content may look better in light mode.
1.Alignment of 3D models between adjacent time points#
Since the coordinate positions of the samples in each period are not fixed, we need to align the samples of multiple periods in three dimensions, so that we can realize the four-dimensional level of spatiotemporal developmental analysis.
Packages#
[1]:
import os
import sys
from pathlib import Path
import numpy as np
import spateo as st
|-----> setting visualization default mode in dynamo. Your customized matplotlib settings might be overritten.
/home/yao/.local/lib/python3.8/site-packages/nxviz/__init__.py:18: UserWarning:
nxviz has a new API! Version 0.7.3 onwards, the old class-based API is being
deprecated in favour of a new API focused on advancing a grammar of network
graphics. If your plotting code depends on the old API, please consider
pinning nxviz at version 0.7.3, as the new API will break your old code.
To check out the new API, please head over to the docs at
https://ericmjl.github.io/nxviz/ to learn more. We hope you enjoy using it!
(This deprecation message will go away in version 1.0.)
warnings.warn(
/home/yao/anaconda3/envs/BGIpy38_tf2/lib/python3.8/site-packages/requests/__init__.py:102: RequestsDependencyWarning: urllib3 (1.26.9) or chardet (5.0.0)/charset_normalizer (2.0.12) doesn't match a supported version!
warnings.warn("urllib3 ({}) or chardet ({})/charset_normalizer ({}) doesn't match a supported "
/home/yao/anaconda3/envs/BGIpy38_tf2/lib/python3.8/site-packages/spaghetti/network.py:36: FutureWarning:
The next major release of pysal/spaghetti (2.0.0) will drop support for all ``libpysal.cg`` geometries. This change is a first step in refactoring ``spaghetti`` that is expected to result in dramatically reduced runtimes for network instantiation and operations. Users currently requiring network and point pattern input as ``libpysal.cg`` geometries should prepare for this simply by converting to ``shapely`` geometries.
Data source#
[3]:
# stage1: E7-9h
stage1_raw_adata = st.sample_data.drosophila(filename="E7-9h_cellbin_tdr_v1.h5ad")
# The embryo pc model and mesh model are reconstructed by ``Three dimensional reconstruction/2.three dims models reconstruction`` using ``E7-9h_cellbin_tdr_v1.h5ad``
stage1_raw_embryo_pc = st.tdr.read_model(filename=f"embryo_raw_pc_model.vtk")
stage1_raw_embryo_mesh = st.tdr.read_model(filename=f"embryo_raw_mesh_model.vtk")
# stage2: E9-10h
stage2_adata = st.sample_data.drosophila(filename="E9-10h_cellbin_tdr_v1.h5ad")
stage2_adata.obsm["before_3d_align_spatial"] = stage2_adata.obsm["tdr_spatial"]
stage2_adata.layers["log1p_X"] = np.log1p(stage2_adata.layers["counts_X"])
Adjust the coordinates of the first model to the final desired coordinates#
[4]:
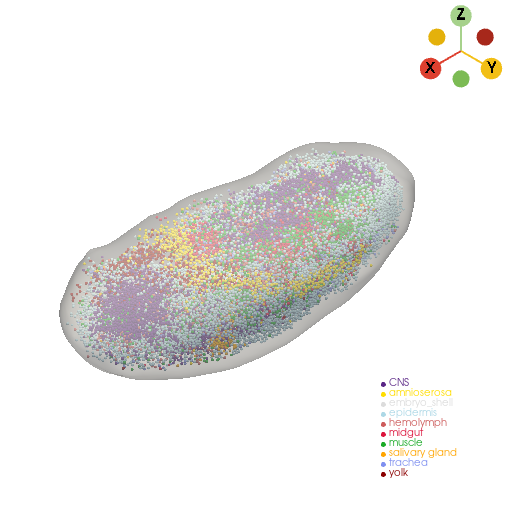
rotate_center = stage1_raw_embryo_pc.center
rotate_angle = (165, 0, -10)
rotate_embryo_pc = st.tdr.rotate_model(model=stage1_raw_embryo_pc, angle=rotate_angle, rotate_center=rotate_center, inplace=False)
rotate_embryo_mesh = st.tdr.rotate_model(model=stage1_raw_embryo_mesh, angle=rotate_angle, rotate_center=rotate_center, inplace=False)
st.pl.three_d_plot(
model=st.tdr.collect_models([rotate_embryo_mesh, rotate_embryo_pc]),
key=["tissue", "tissue"],
model_style=["surface", "points"],
model_size=[0, 3],
jupyter="static",
)
[5]:
stage1_adata = stage1_raw_adata[np.asarray(rotate_embryo_pc.point_data["obs_index"]), :].copy()
stage1_adata.layers["log1p_X"] = np.log1p(stage1_adata.layers["counts_X"])
# stage1_adata.uns["pp"] = {}
# dyn.pp.normalize_cell_expr_by_size_factors(adata=stage1_adata, layers="normalize_X", X_total_layers=True)
stage1_adata.obsm["before_3d_align_spatial"] = np.asarray(rotate_embryo_pc.points)
stage1_adata
[5]:
AnnData object with n_obs × n_vars = 25857 × 8136
obs: 'area', 'slices', 'nGenes', 'nCounts', 'pMito', 'nMito', 'pass_basic_filter', 'scc', 'auto_anno', 'anno_cell_type', 'anno_tissue', 'anno_germ_layer', 'actual_stage'
uns: 'PCs', '__type', 'auto_anno_result', 'dendrogram_anno_cell_type', 'dendrogram_anno_germ_layer', 'dendrogram_anno_tissue', 'explained_variance_ratio_', 'neighbors', 'pca_mean', 'pca_valid_ind', 'pearson_X_neighbors', 'rank_genes_groups', 'rank_genes_groups_anno_cell_type', 'rank_genes_groups_anno_germ_layer', 'rank_genes_groups_anno_tissue', 'scc', 'spatial'
obsm: 'align_spatial', 'bbox', 'contour', 'pearson_X_pca', 'pearson_X_umap', 'spatial', 'tdr_spatial', 'before_3d_align_spatial'
layers: 'counts_X', 'log1p_X', 'pearson_X', 'spliced', 'unspliced'
Alignment of 3D models of two stages#
[6]:
align_samples, align_samples_ref = st.tl.models_align_ref(
models=[stage1_adata, stage2_adata],
models_ref=None,
n_sampling=2000,
sampling_method="trn",
layer="log1p_X",
spatial_key="before_3d_align_spatial",
key_added="3d_align_spatial",
device="0",
)
align_samples
|-----> [Running TRN] in progress: 100.0000%
|-----> [Running TRN] finished [378.3584s]
|-----> [Running TRN] in progress: 100.0000%
|-----> [Running TRN] finished [381.4986s]
|-----> [Models alignment] in progress: 100.0000%|-----------> Filtered all samples for common genes. There are 7890 common genes.
|-----> [Models alignment] in progress: 100.0000%
|-----> [Models alignment] finished [18.6270s]
[6]:
[AnnData object with n_obs × n_vars = 25857 × 8136
obs: 'area', 'slices', 'nGenes', 'nCounts', 'pMito', 'nMito', 'pass_basic_filter', 'scc', 'auto_anno', 'anno_cell_type', 'anno_tissue', 'anno_germ_layer', 'actual_stage'
uns: 'PCs', '__type', 'auto_anno_result', 'dendrogram_anno_cell_type', 'dendrogram_anno_germ_layer', 'dendrogram_anno_tissue', 'explained_variance_ratio_', 'neighbors', 'pca_mean', 'pca_valid_ind', 'pearson_X_neighbors', 'rank_genes_groups', 'rank_genes_groups_anno_cell_type', 'rank_genes_groups_anno_germ_layer', 'rank_genes_groups_anno_tissue', 'scc', 'spatial', 'latter_models_align'
obsm: 'align_spatial', 'bbox', 'contour', 'pearson_X_pca', 'pearson_X_umap', 'spatial', 'tdr_spatial', 'before_3d_align_spatial', '3d_align_spatial'
layers: 'counts_X', 'log1p_X', 'pearson_X', 'spliced', 'unspliced',
AnnData object with n_obs × n_vars = 24304 × 8484
obs: 'area', 'slices', 'x', 'y', 'z', 'nGenes', 'nCounts', 'pMito', 'nMito', 'pass_basic_filter', 'scc', 'auto_anno', 'anno_cell_type', 'anno_tissue', 'anno_germ_layer', 'actual_stage'
uns: 'PCs', '__type', 'auto_anno_result', 'dendrogram_anno_cell_type', 'dendrogram_anno_germ_layer', 'dendrogram_anno_tissue', 'dendrogram_scc', 'explained_variance_ratio_', 'neighbors', 'pca_mean', 'pca_valid_ind', 'pearson_X_neighbors', 'rank_genes_groups', 'rank_genes_groups_anno_cell_type', 'rank_genes_groups_anno_germ_layer', 'rank_genes_groups_anno_tissue', 'scc', 'scc_colors', 'spatial', 'former_models_align'
obsm: 'align_spatial', 'bbox', 'contour', 'pearson_X_pca', 'pearson_X_umap', 'spatial', 'tdr_spatial', 'before_3d_align_spatial', '3d_align_spatial'
layers: 'counts_X', 'log1p_X', 'pearson_X', 'spliced', 'unspliced']
[7]:
stage1_aligned_adata_ref = align_samples_ref[0].copy()
stage2_aligned_adata_ref = align_samples_ref[1].copy()
stage1_aligned_adata = align_samples[0].copy()
stage2_aligned_adata = align_samples[1].copy()
Generate aligned 3D models#
stage1 point cloud model#
[8]:
stage1_tissue_colormap = {
"CNS": "#5A2686",
"midgut": "#DC143C",
"amnioserosa": "#FFDD00",
"salivary gland": "#FFA500",
"epidermis": "#ADD8E6",
"muscle": "#1AAB27",
"trachea": "#7F90F0",
"hemolymph": "#CD5C5C",
"yolk": "#8B0000",
}
stage1_raw_ref_pc = st.tdr.construct_pc(
adata=stage1_aligned_adata_ref.copy(),
spatial_key="before_3d_align_spatial",
groupby="anno_tissue",
key_added="tissue",
colormap=stage1_tissue_colormap,
)
stage1_aligned_ref_pc = st.tdr.construct_pc(
adata=stage1_aligned_adata_ref.copy(),
spatial_key="3d_align_spatial",
groupby="anno_tissue",
key_added="tissue",
colormap=stage1_tissue_colormap,
)
stage1_raw_pc = st.tdr.construct_pc(
adata=stage1_aligned_adata.copy(),
spatial_key="before_3d_align_spatial",
groupby="anno_tissue",
key_added="tissue",
colormap=stage1_tissue_colormap,
)
stage1_aligned_pc = st.tdr.construct_pc(
adata=stage1_aligned_adata.copy(),
spatial_key="3d_align_spatial",
groupby="anno_tissue",
key_added="tissue",
colormap=stage1_tissue_colormap,
)
stage2 point cloud model#
[9]:
stage2_tissue_colormap = {
"CNS": "#5A2686",
"midgut": "#DC143C",
"amnioserosa": "#FFDD00",
"salivary gland": "#FFA500",
"epidermis": "#ADD8E6",
"muscle": "#1AAB27",
"trachea": "#7F90F0",
"hemolymph": "#CD5C5C",
"yolk": "#8B0000",
}
stage2_raw_ref_pc = st.tdr.construct_pc(
adata=stage2_aligned_adata_ref.copy(),
spatial_key="before_3d_align_spatial",
groupby="anno_tissue",
key_added="tissue",
colormap=stage2_tissue_colormap,
)
stage2_aligned_ref_pc = st.tdr.construct_pc(
adata=stage2_aligned_adata_ref.copy(),
spatial_key="3d_align_spatial",
groupby="anno_tissue",
key_added="tissue",
colormap=stage2_tissue_colormap,
)
stage2_raw_pc = st.tdr.construct_pc(
adata=stage2_aligned_adata.copy(),
spatial_key="before_3d_align_spatial",
groupby="anno_tissue",
key_added="tissue",
colormap=stage2_tissue_colormap,
)
stage2_aligned_pc = st.tdr.construct_pc(
adata=stage2_aligned_adata.copy(),
spatial_key="3d_align_spatial",
groupby="anno_tissue",
key_added="tissue",
colormap=stage2_tissue_colormap,
)
Check that the alignment results at the overall level#
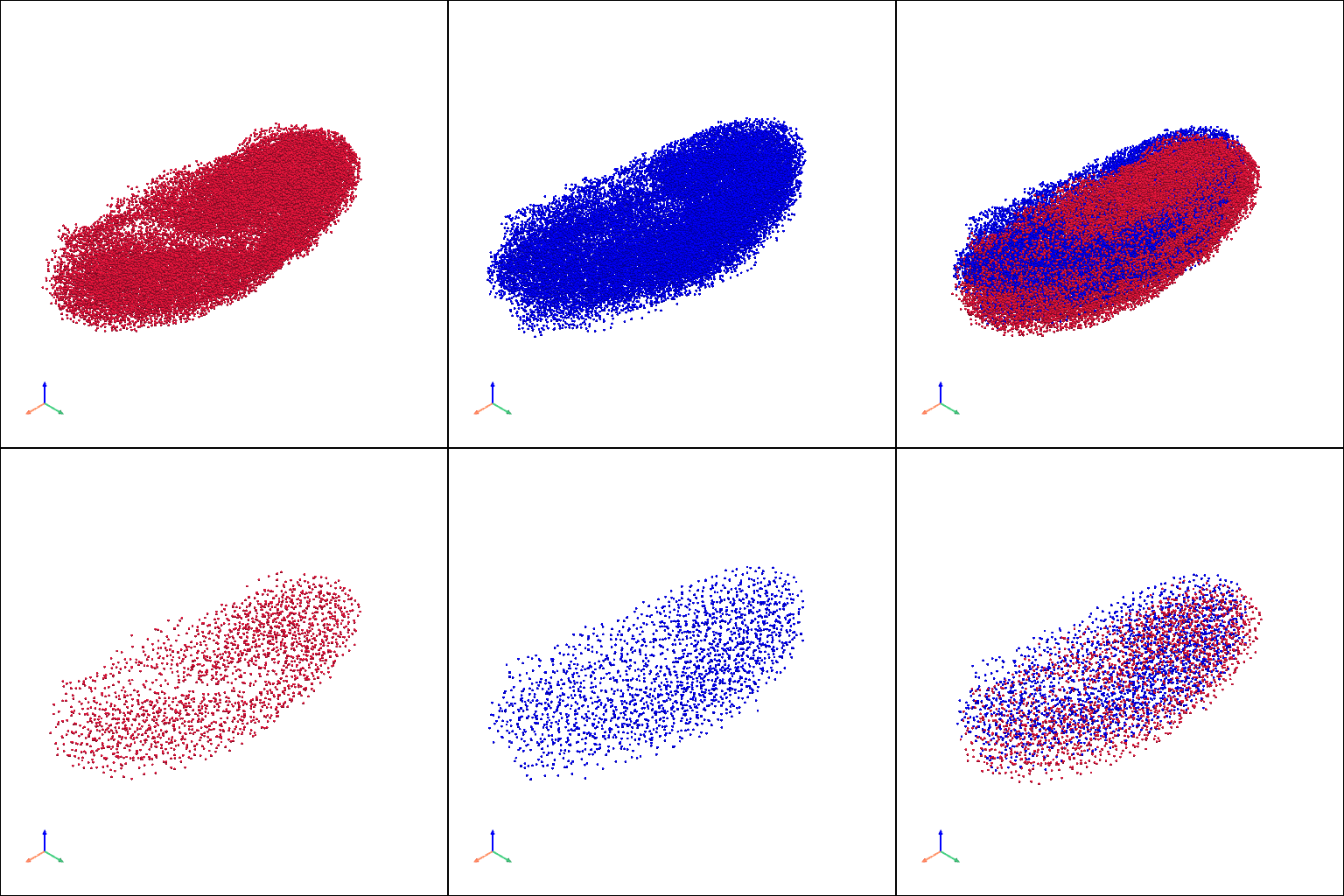
raw embryo model#
[11]:
raw_pcs_v = [
stage1_raw_pc.copy(), stage2_raw_pc.copy(), st.tdr.collect_models([stage1_raw_pc.copy(), stage2_raw_pc.copy()]),
stage1_raw_ref_pc.copy(), stage2_raw_ref_pc.copy(), st.tdr.collect_models([stage1_raw_ref_pc.copy(), stage2_raw_ref_pc.copy()])
]
st.pl.three_d_multi_plot(
model=st.tdr.collect_models(raw_pcs_v),
colormap=["#DC143C", "#0000FF", ["#DC143C", "#0000FF"], "#DC143C", "#0000FF", ["#DC143C", "#0000FF"]],
model_style="points",
model_size=3,
off_screen=False,
shape=(2, 3),
window_size=(512*3, 512*2),
show_legend=False,
jupyter="static",
)
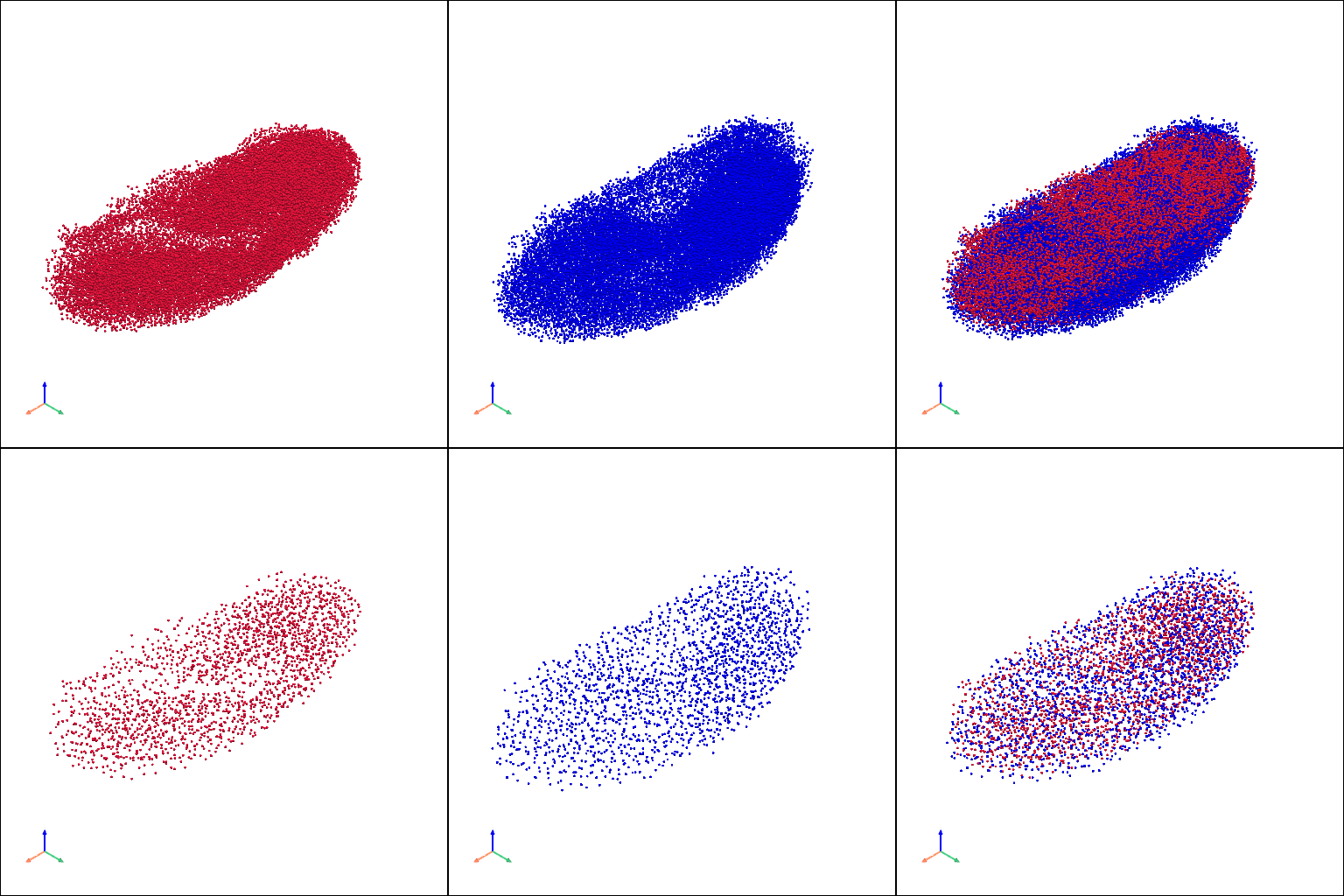
aligned embryo model#
[12]:
aligned_pcs_v = [
stage1_aligned_pc.copy(), stage2_aligned_pc.copy(), st.tdr.collect_models([stage1_aligned_pc.copy(), stage2_aligned_pc.copy()]),
stage1_aligned_ref_pc.copy(), stage2_aligned_ref_pc.copy(), st.tdr.collect_models([stage1_aligned_ref_pc.copy(), stage2_aligned_ref_pc.copy()])
]
st.pl.three_d_multi_plot(
model=st.tdr.collect_models(aligned_pcs_v),
colormap=["#DC143C", "#0000FF", ["#DC143C", "#0000FF"], "#DC143C", "#0000FF", ["#DC143C", "#0000FF"]],
model_style="points",
model_size=3,
off_screen=False,
shape=(2, 3),
window_size=(512*3, 512*2),
show_legend=False,
jupyter="static",
)
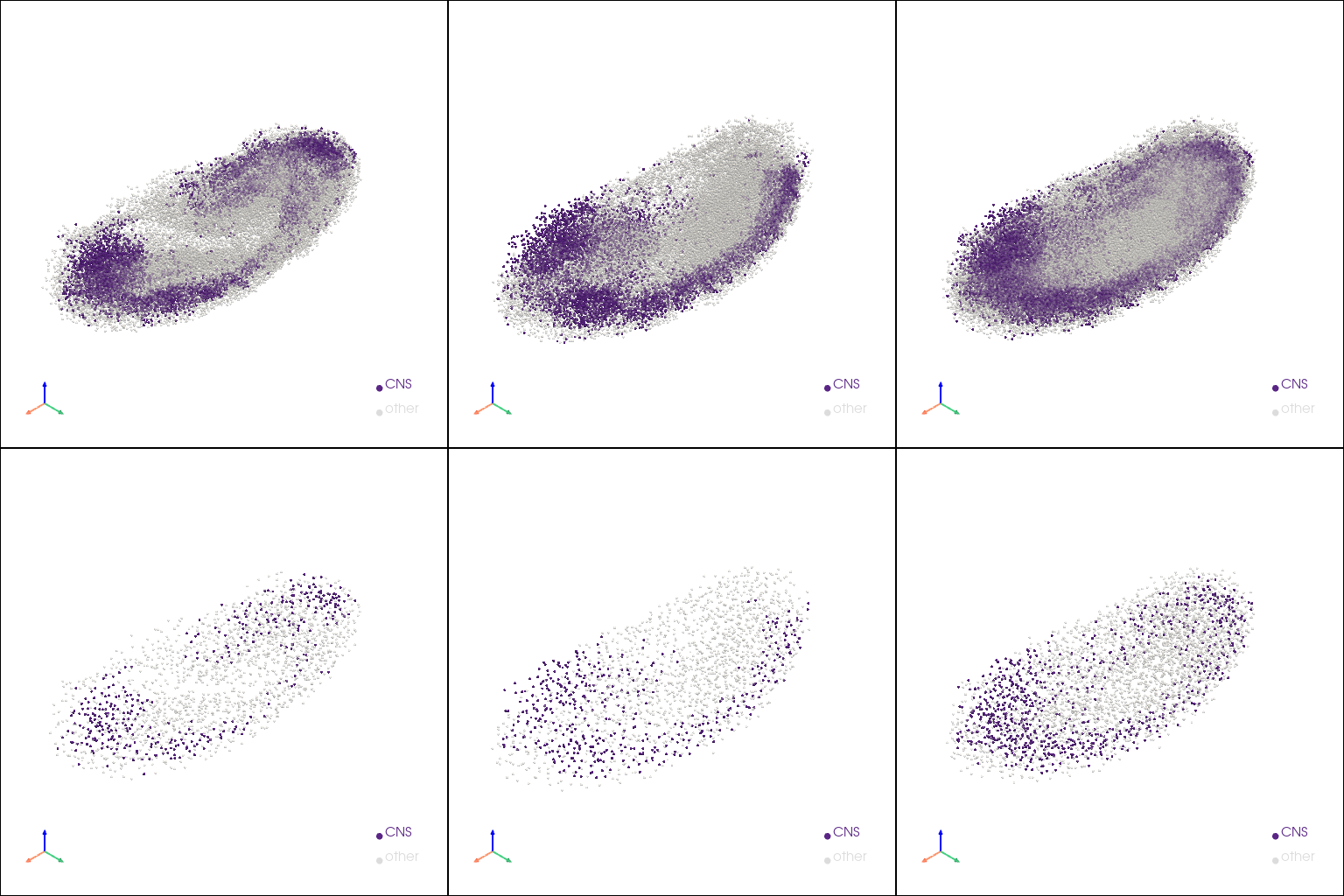
aligned embryo models with CNS#
[13]:
aligned_pcs_v = []
for pcs in [[stage1_aligned_pc.copy(), stage2_aligned_pc.copy()], [stage1_aligned_ref_pc.copy(), stage2_aligned_ref_pc.copy()]]:
for pc in pcs:
cns_label = np.asarray(pc.point_data["tissue"])
cns_label[cns_label != "CNS"] = "other"
st.tdr.add_model_labels(model=pc, labels=cns_label, key_added="check_alignment", where="point_data", inplace=True,
colormap={"CNS": "#5A2686", "other": "gainsboro"}, alphamap={"CNS": 1.0, "other": 0.5})
pcs.append(st.tdr.collect_models(pcs.copy()))
aligned_pcs_v.extend(pcs)
st.pl.three_d_multi_plot(
model=st.tdr.collect_models(aligned_pcs_v),
key="check_alignment",
model_style="points",
model_size=3,
off_screen=False,
shape=(2, 3),
window_size=(512*3, 512*2),
jupyter="static",
)
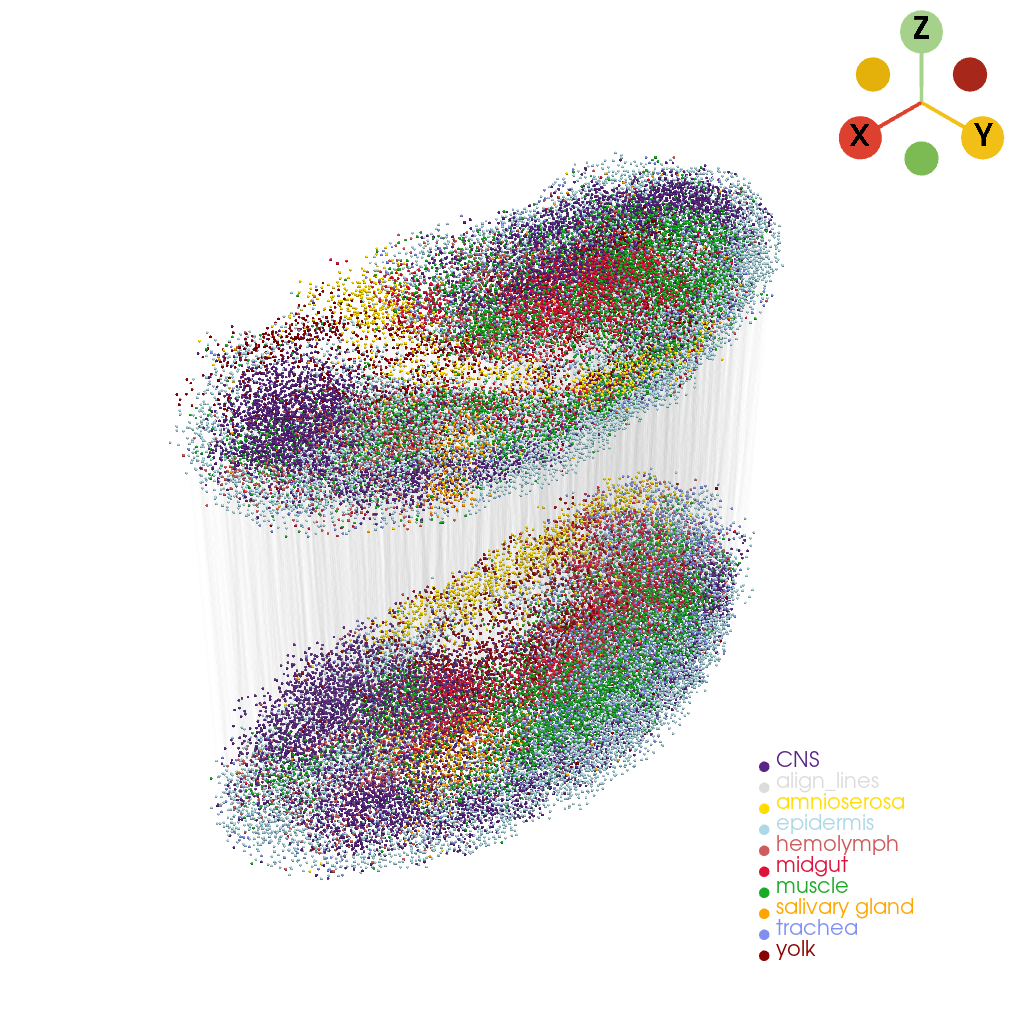
Check that the alignment results at the cell level#
[14]:
cells_align_dict = stage1_aligned_adata.uns["latter_models_align"]
align_lines = st.tdr.construct_align_lines(
model1_points=cells_align_dict["mapping_X"].copy(),
model2_points=cells_align_dict["mapping_Y"].copy() + np.asarray([0, 0, - 300]),
key_added="check_align", label="align_lines", color="gainsboro")
[15]:
stage1_aligned_pc_v = stage1_aligned_pc.copy()
stage2_aligned_pc_v = stage2_aligned_pc.copy()
stage2_aligned_pc_v.points[:, 2] = stage2_aligned_pc_v.points[:, 2] - 300
[16]:
st.pl.three_d_plot(
model=st.tdr.collect_models([align_lines, stage1_aligned_pc_v, stage2_aligned_pc_v]),
key=["check_align", "tissue", "tissue"],
opacity=[0.01, 1.0, 1.0],
model_style=["wireframe", "points", "points"],
model_size=3,
off_screen=False,
window_size=(1024, 1024),
jupyter="static",
)
[ ]: